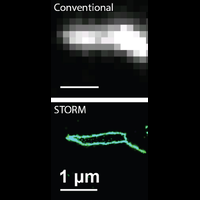
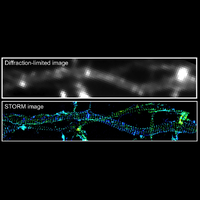
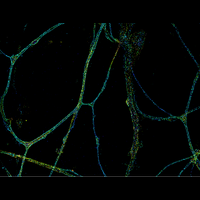
Understanding the mechanisms of cellular function and their dysfunction in disease requires a detailed picture of the molecular interactions in cells. In particular, we need imaging tools with single-molecule sensitivity, molecular-scale resolution, and dynamic imaging capability to allow direct visualization of molecular interactions in cells, as well as tools that can simultaneously image large numbers of genes, ideally at the genome scale, to probe how collective actions of these molecules give rise to cellular and tissue functions. The research in the Zhuang laboratory is aimed at developing such imaging methods and applying them to problems of biomedical interest.
Students and postdoctoral fellows in the Zhuang laboratory apply their diverse backgrounds in chemistry, physics, biology, and engineering to develop novel imaging methods, molecular probes, and image analysis algorithms, and to exploit these tools to study a variety of interesting biological problems, ranging from the structure of chromatin and chromosomes and the regulation of gene expression to sub-cellular structures in neurons and neuronal connectivity. Our current research is focused in three major areas: (1) Genome-scale imaging and single-cell spatial genomics, (2) super-resolution imaging, and (3) single-molecule biology.
Multi-cellular tissue function arises from integration of genetically identical cells that display distinct behaviors, which are in turn determined by their differential gene expression. Thus, measurements of the transcriptomes of individual cells are critical for our understanding of the molecular origin of cell behavior and tissue function. These measurements will allow us to address fundamental questions in a wide range of topics, from the mechanisms of gene regulation to the development of cell fate and the organization of distinct cell types in tissues. Indeed, recent advances in single-cell transcriptomics, primarily driven by advances in next generation sequencing, has begun to transform many areas of biology. However, these approaches typically require dissociation of cells from tissues and extraction of RNAs from cells; hence the intracellular locations of RNAs and the spatial context of the cells, which are critical for cellular and tissue functions, are lost. In order to fully characterize the transcriptome, we need to determine not only the copy number of each RNA species within individual cells but also the spatial context of these RNAs.
Recently, we developed a massively multiplexed single-molecule imaging method, termed multiplexed error robust fluorescence in situ hybridization (MERFISH), that is capable of imaging many RNA species in individual cells. In this approach, we label RNA species in a combinatorial manner using fluorescence in situ hybridization (FISH) and error-robust barcodes, and read out these barcodes using sequential rounds of imaging with single-molecule resolution. Our initial MERFISH experiments demonstrated the ability to image ~1000 RNA species in individual cells, determining both the copy numbers and spatial distributions of these RNAs. These experiments revealed distinct sub-cellular distributions of RNAs that correlate with the properties of their encoded proteins and provided new insights into gene regulatory networks. We are currently advancing our MERFISH imaging capabilities to increase both the number of RNA species and the number of cells that can be measured in each experiment. We are also applying this method to determine the spatial distribution of RNAs inside neurons and the spatial organization of transcriptionally distinct cell types in brain tissues and tumors.
We have also developed a multiplexed FISH method that allows us to image numerous genomic loci and trace the conformation of individual chromosomes in single cells. The genome is folded into an intricate 3D structure in the nucleus, and this 3D structure is critical for the regulation of gene expression and other genome functions. However, many gaps remain in our understanding of the 3D organization of chromosomes. Our multiplexed FISH method has provided novel insights into the spatial organization of chromatin domains and compartments in individual chromosomes. We are currently developing the ability to trace the 3D paths of whole chromosomes at nanometer-scale resolution and using this approach to study the spatial organization of the genome and its role in various genome functions.
In addition to transcriptional regulation, translation is also under tight regulation to ensure protein production in the right time and place in cells. We have also been developing imaging methods that allow translation activities on individual mRNA molecules to be imaged in real time in live cells.
Selected publications:
R. A. Saunders, W. E. Allen, X. Pan, J. Sandhu, J. Lu, T. K. Lau, K. Smolyar, Z. A. Sullivan, C. Dulac, J. S. Weissman, X. Zhuang
S. Liu, C. Y. Wang, P. Zheng, B. B. Jia, N. R. Zemke, P. Ren, H. L. Park, B. Ren, X. Zhuang
M. Zhang, X. Pan, W. Jung, A. Halpern, S. W. Eichhorn, Z. Lei, L. Cohen, K. A. Smith, B. Tasic, Z. Yao, H. Zeng, X. Zhuang
W. E. Allen, T. R. Blosser, Z. A. Sullivan, C. Dulac, X. Zhuang
T. Lu, C. E. Ang, X. Zhuang
R. Fang, C. Xia, J. L. Close, M. Zhang, J. He, Z. Huang, A. R. Halpern, B. Long, J. A. Miller, E. S. Lein, X. Zhuang
M. Zhang, S. W. Eichhorn, B. Zingg, Z. Yao, K. Cotter, H. Zeng, H. Dong, X. Zhuang
R. Chen, T. R. Blosser, M. N. Djekidel, J. Hao, A. Bhattacherjee, W. Chen, L. M. Tuesta, X. Zhuang, Y. Zhang
J. -H. Su, P. Zheng, S. S. Kinrot, B. Bintu, X. Zhuang
C. Xia, J. Fan, G. Emanuel, J. Hao, X. Zhuang
C. Wang, T. Lu, G. Emanuel, H.P. Babcock, X. Zhuang
J. Moffitt, D. Bambah-Mukku, S. Eichhorn, E. Vaughn, K. Shekhar, N. Rubinstein, J. Hao, A. Regev, C. Dulac, X. Zhuang
B. Bintu, L. Mateo, J. Su, N. Sinnott-Armstrong, M. Parker, S. Kinrot, K. Yamaya, A. Boettiger, X. Zhuang
G. Emanuel, J.R. Moffitt, X. Zhuang
J.R. Moffitt, J. Hao, D. Bambah-Mukkua, T. Lu, C. Dulac, X. Zhuang
J.R. Moffitt, J. Hao, G. Wang, K.H. Chen, H.P. Babcock, X. Zhuang
S. Wang, J.H. Su, B.J. Beliveau, J.R. Moffitt, C.T. Wu, X. Zhuang
K.H. Chen, A.N. Boettiger, J.R. Moffitt, S. Wang, X. Zhuang
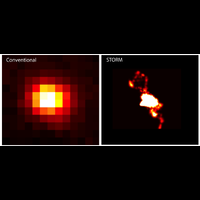
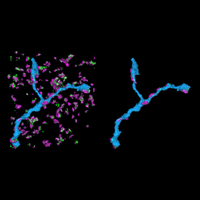
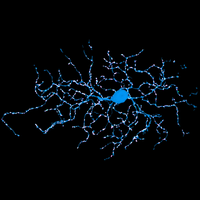
Fluorescence microscopy is one of the most widely used imaging methods in biomedical research. Its molecular/chemical specificity and live-cell compatibility have made fluorescence microscopy a particularly powerful tool for biological research, and numerous breakthroughs in biology have been enabled by this imaging modality. However, the spatial resolution of light microscopy, classically limited by the diffraction of light to several hundred nanometers, is substantially larger than the typical molecular-length scales in cells, preventing a detailed characterization of most subcellular structures.
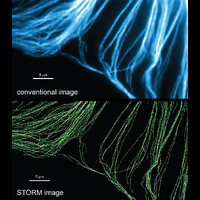
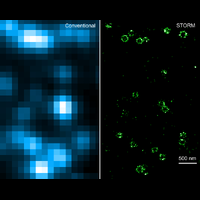
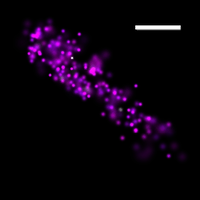
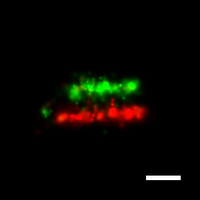
To overcome this limit, we have developed a single-molecule-based super-resolution light microscopy method, stochastic optical reconstruction microscopy (STORM). In this method, we introduced the use of photoswitchable fluorescent probes to temporally separate the spatially overlapping images of individual molecules, allowing the positions of these molecules to be precisely determined and super-resolution images to be reconstructed from their molecular coordinates. Using STORM, we have achieved three-dimensional, multicolor fluorescence imaging of cells and tissues with sub-diffraction-limit resolution. Through innovations in optical physics, molecular probes and labeling chemistry, and analysis algorithms, we have obtained a resolution of sub-10 nm, demonstrated live-cell STORM imaging with sub-second time resolution, and extended the STORM capabilities to image large tissue volumes. We are currently working on advancing super-resolution imaging by further increasing its spatial and temporal resolution and improving its deep-tissue, live-animal imaging capabilities.
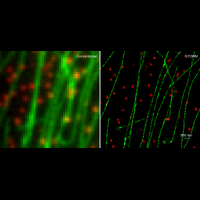
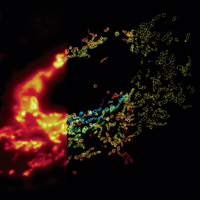
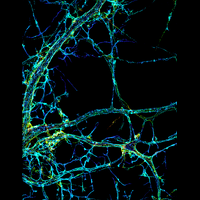
We have applied STORM to a variety of problems in cell biology and neurobiology. These studies have elucidated the high-resolution structures of many molecular assemblies and led to the discovery of previously unknown cellular structures in cells. For example, we discovered a periodic, membrane-associated, cytoskeleton structure in neurons made of actin, spectrin, and associated molecules, which ubiquitously exists in diverse neuronal cell types and in a wide variety of animal species, ranging from C. elegans to H. sapiens. Our studies have also provided novel insights into the three-dimensional spatial organization of chromatin and chromosomes in the nucleus, the molecular architectures of synapses, the spatial distributions and molecular identities of synapse on neurons, etc. Currently, we are focusing on the following areas of biological investigation: 1) sub-cellular structures, in particular the membrane associated periodic cytoskeleton structure in the neurons, and 2) the three-dimensional structures and dynamics of chromatin and chromosomes in the nucleus.
STORM Image Gallery
Click here for a more detailed description of recent research in the lab.
STORM Workshop (August 2010, April 2012)
Selected publications:
E. Heller, N. Kurup, X. Zhuang
R. Zhou, B. Han, R. Nowak, Y. Lu, E. Heller, C. Xia, A. H. Chishti, V. M. Fowler, X. Zhuang
R. Zhou, B. Han, C. Xia, X. Zhuang
G. Wang, D.J. Simon, Z. Wu, D.M. Belsky, E. Heller, M.K. O'Rourke, N.T. Hertz, H. Molina, G. Zhong, M. Tessier-Lavigne, X. Zhuang
A.N. Boettiger, B. Bintu, J.R. Moffitt, S. Wang, B.J. Beliveau, G. Fudenberg, M. Imakaev, L.A. Mirny, C.T. Wu, X. Zhuang
J.B. French, S.A. Jones, H. Deng, A.M. Pedley, D. Kim, C.Y. Chan, H. Hu, R.J. Pugh, H. Zhao, Y. Zhang, T.J. Huang, Y. Fang, X. Zhuang, S.J. Benkovic
J. He, R. Zhou, Z. Wu, M.A. Carrasco, P.T. Kurshan, J.E. Farley, D.J. Simon, G. Wang, B. Han, J. Hao, E. Heller, M.R. Freeman, K. Shen, T. Maniatis, M. Tessier-Lavigne, X. Zhuang
Y.M. Sigal, C.M. Speer, H.P. Babcock, X. Zhuang
G. Zhong, J. He, R. Zhou, D. Lorenzo, H.P. Babcock, V. Bennett, X. Zhuang
J. Chung, S. Shim, R. Everley, S. Gygi, X. Zhuang, D. Clapham
K. Xu, G. Zhong, X. Zhuang
Y. Doksani, J. Wu, T. de Lange, X. Zhuang
J. Vaughan, S. Jia, X. Zhuang
S. Jones, S-H. Shim, J. He, X. Zhuang
A. Dani, B. Huang, J. Bergan, C. Dulac, X. Zhuang
B. Huang, W. Wang, M. Bates, X. Zhuang
M. Bates, B. Huang, G. T. Dempsey, X. Zhuang
M. J. Rust, M. Bates, X. Zhuang
We have developed and applied single-molecule fluorescence imaging and spectroscopy techniques to study complex biomolecular systems in vitro. In particular, we use the highly sensitive distance dependence of fluorescence resonance energy transfer (FRET) to detect structural changes at the single-molecule level. We are also developing novel single-molecule methods to detect the motion dynamics within biomolecular complexes.
An area of particular interest to us is the interactions of proteins with nucleic acids. Many essential cellular reactions, such as DNA replication, transcription, RNA processing, and protein synthesis, involve DNA-protein or RNA-protein complexes. Understanding nucleic acid-protein interactions is thus crucial for deciphering the molecular mechanisms underlying many important biological processes. Using single-molecule methods, we directly visualize the assembly and function of these molecular complexes in real time. These experiments allow us to observe transient states that are difficult to detect by classical ensemble experiments, to probe the dynamic interactions between DNA, RNA and proteins, and to determine the relationship between the structural dynamics and function for these molecular complexes. Using this approach, we have studied the assembly process, catalytic cycle, and structure-function relationship of several nucleic acid-interacting enzymes, including ribozymes, telomerase, HIV reverse transcriptase and chromatin-remodeling enzymes.
Our current focus is on ATP-dependent chromatin remodeling enzymes. Our single-molecule studies have revealed the translocation step sizes of ISWI-family and SWI/SNF-family remodeling enzymes and elucidated how DNA is translocated across the nucleosome by these enzymes. We also elucidated the molecular mechanism by which the linker DNA length and histone modifications regulate ISWI-mediated nucleosome translocation to generate evenly spaced nucleosomes in heterochromatin. We are continuing to study these enzymes, how they remodel chromatin structures, and how the remodeling process is regulated.
Click here for a more detailed description of recent research in the lab.
Selected publications:
P. Kosuri, B.D. Altheimer, M. Dai, P. Yin, X. Zhuang
C. Wang, B. Han, R. Zhou, X. Zhuang
W.L. Hwang, S. Deindl, B.T. Harada, X. Zhuang
S. Deindl, W.L. Hwang, S.K. Hota, T.R. Blosser, P. Prasad, B. Bartholomew, X. Zhuang
T. Blosser, J. Yang, M. Stone, G. Narlikar, X. Zhuang
S. Liu, E. Abbondanzieri, J. W. Rausch, S. F. J. Le Grice, X. Zhuang
E. Abbondanzieri, G. Bokinsky, J. W. Rausch, J. X. Zhang, S. F. J. Le Grice, X. Zhuang
M. D. Stone, M. Mihalusova, C. M. O'Connor, R. Prathapam, K. Collins, X. Zhuang